糖尿病遗传风险检测挑战赛 - Coggle 30 Days of ML(22年7月)
本文最后更新于:2022年9月13日 07:56
任务1:报名比赛
步骤1:报名比赛http://challenge.xfyun.cn/topic/info?type=diabetes&ch=ds22-dw-zmt05

步骤2:下载比赛数据(点击比赛页面的赛题数据)
步骤3:解压比赛数据,并使用pandas进行读取
1
2
3
4
5
6import pandas as pd
train_df = pd.read_csv('./比赛训练集.csv', encoding='gbk')
test_df = pd.read_csv('./比赛测试集.csv', encoding='gbk')
print(train_df.shape,test_df.shape)
步骤4:查看训练集和测试集字段类型
1
2train_df.dtypes
test_df.dtypes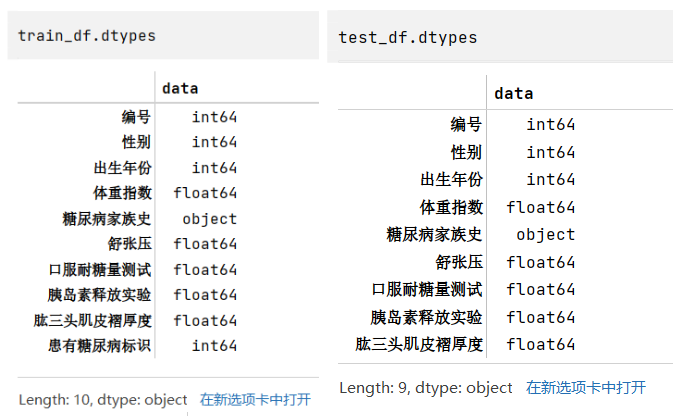
任务2:比赛数据分析
步骤1:统计字段的缺失值,计算缺失比例;
- 通过缺失值统计,训练集和测试集的缺失值分布是否一致?
- 通过缺失值统计,有没有缺失比例很高的列?
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19train_nan_df = pd.DataFrame(columns=['标签','训练集缺失个数','训练集缺失比例'])
i = 0
for tag in list(train_df.columns):
num = train_df[tag].isnull().sum()
num_rate = num/len(train_df)
train_nan_df.loc[i] = [tag, num, num_rate]
i += 1
# train_nan_df
test_nan_df = pd.DataFrame(columns=['标签','测试集缺失个数','测试集缺失比例'])
i = 0
for tag in list(test_df.columns):
num = test_df[tag].isnull().sum()
num_rate = num/len(test_df)
test_nan_df.loc[i] = [tag, num, num_rate]
i += 1
# test_nan_df
train_nan_df.merge(test_nan_df,how='left')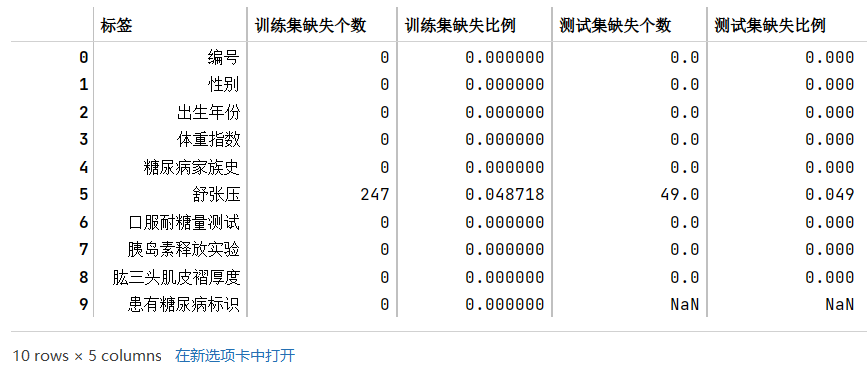
根据结果可知,训练集和测试集的缺失值分布一致,都是有且只有舒张压这一个标签含缺失值,且比例都在4.8-4.9%左右,缺失的比例其实也不算是特别高。
步骤2:分析字段的类型
- 有多少数值类型、类别类型?
- 你是判断字段类型的?
1
train_df.head() # tail()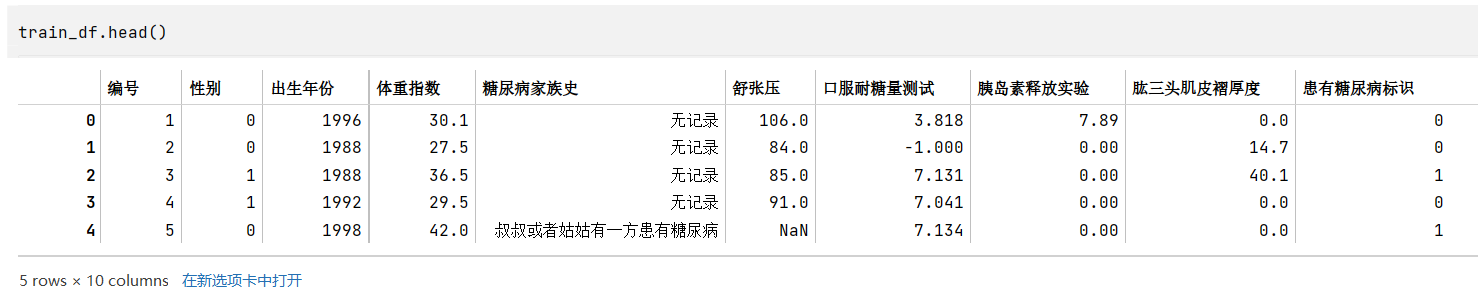
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18train_category_num_df = pd.DataFrame(columns=['标签','训练集该标签不同值个数'])
i = 0
for tag in list(train_df.columns):
num = len(train_df[tag].value_counts())
train_category_num_df.loc[i] = [tag, num]
i += 1
# train_category_num_df
test_category_num_df = pd.DataFrame(columns=['标签','测试集该标签不同值个数'])
i = 0
for tag in list(test_df.columns):
num = len(test_df[tag].value_counts())
test_category_num_df.loc[i] = [tag, num]
i += 1
# test_category_num_df
train_category_num_df.merge(test_category_num_df,how='left')
# 列包含NaN,因此dtype必须升级为浮点dtype以容纳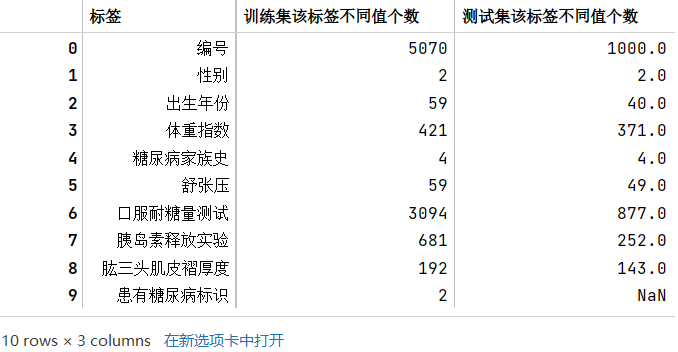
字段类型的判断主要通过根据常识(如性别)、观察原始数据(如糖尿病家族史)、以及各个标签的类型数量(如数量较多的可能就是连续性数据,是数值类型)来判断
- 数值类型:编号(和时序无关,后期没必要研究),出生年份,体重指数,舒张压,口服耐糖量测试,胰岛素释放实验,肱三头肌皮褶厚度
- 类别类型:性别(两类),糖尿病家族史(类型),患有糖尿病标识
- 当然,后期为了改善模型效果,也可以将一些连续性数据转换成类别,比如体重指数可以根据国家标准划分胖瘦类型,成为类别类型
- 其中,糖尿病家族史有四类,但是查看其类型,和“叔叔或者姑姑有一方患有糖尿病”应是同一表述,但是被分到两类,应该合并成一类为“叔叔或姑姑有一方患有糖尿病”
1
2
3
4
5
6
7
8train_df.糖尿病家族史.value_counts()
train_df.loc[train_df.糖尿病家族史 == '叔叔或者姑姑有一方患有糖尿病','糖尿病家族史'] = '叔叔或姑姑有一方患有糖尿病'
train_df.糖尿病家族史.value_counts()
train_df['糖尿病家族史'] = train_df['糖尿病家族史'].astype('int64')
test_df['糖尿病家族史'] = test_df['糖尿病家族史'].astype('int64')
train_df.dtypes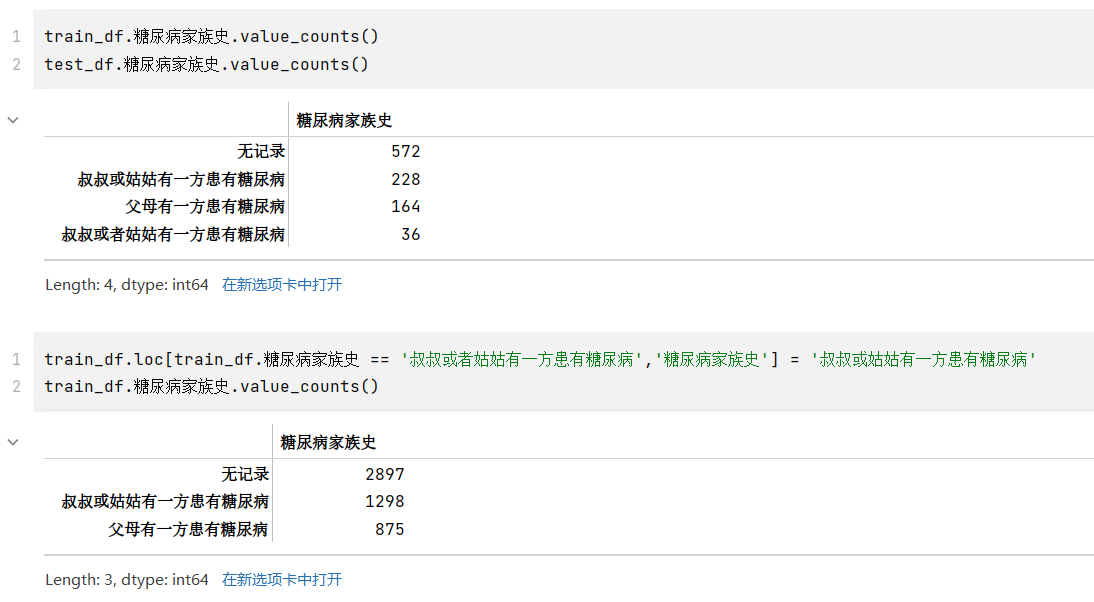
步骤3:计算字段相关性
- 通过
.corr()计算字段之间的相关性 - 有哪些字段与标签的相关性最高?
- 尝试使用其他可视化方法将字段 与 标签的分布差异进行可视化
1
2
3
4tag_list = train_df.columns.tolist()
tag_list.pop(tag_list.index("编号"))
corr = train_df[tag_list].corr()
corr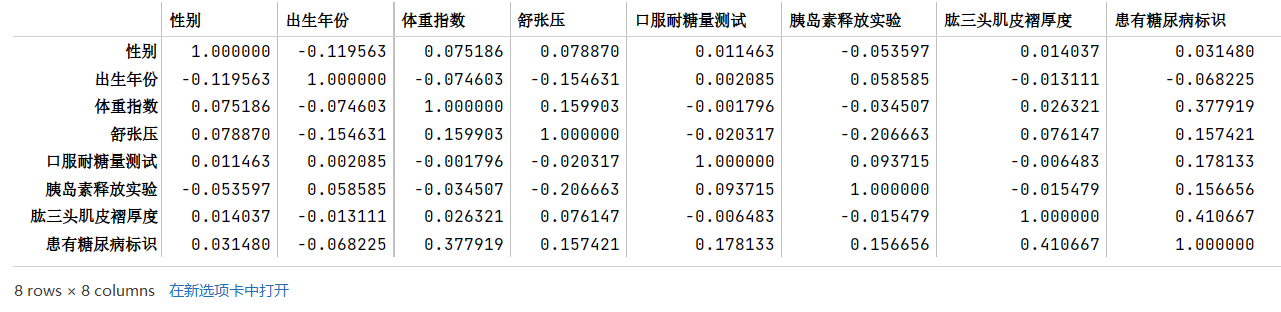
虽然获得了相关系数矩阵,但是不便于分析结果,将其进行可视化,使用seaborn绘制热力图
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21from string import ascii_letters
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
sns.set_theme(style="white")
plt.rcParams['font.sans-serif'] = ['SimHei'] # 黑体
plt.rcParams['axes.unicode_minus']=False # 负号
# Generate a mask for the upper triangle
mask = np.triu(np.ones_like(corr, dtype=bool))
# Set up the matplotlib figure
f, ax = plt.subplots(figsize=(8, 8))
# Generate a custom diverging colormap
cmap = sns.diverging_palette(230, 20, as_cmap=True)
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(corr, mask=mask, cmap=cmap, vmax=0.5, center=0, vmin=-0.3,
square=True, linewidths=2, cbar_kws={"shrink": .5})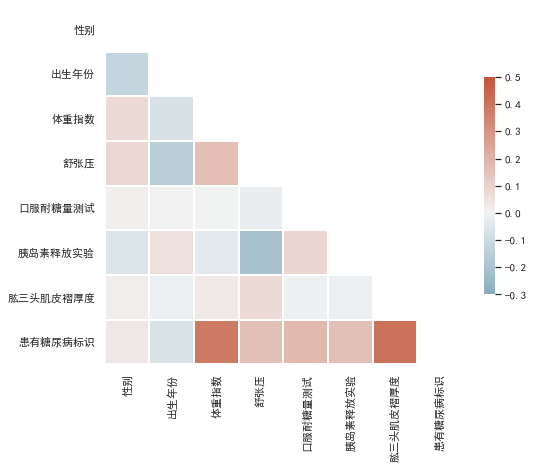
颜色越深,表示和标签的相关性最高,也就是可以重点关注一下体重指数和肱三头肌褶厚度。
同时,还对不同的自变量之间进行相关性分析,按照是否有患有糖尿病标识绘制多变量联合分布pairplot图
1
2
3
4
5
6sns.set_theme(style="ticks")
plt.rcParams['font.sans-serif'] = ['SimHei'] # 黑体
plt.rcParams['axes.unicode_minus']=False # 负号
sns.pairplot(train_df.drop(columns=['编号','性别']), hue='患有糖尿病标识',plot_kws={'alpha': 0.5})
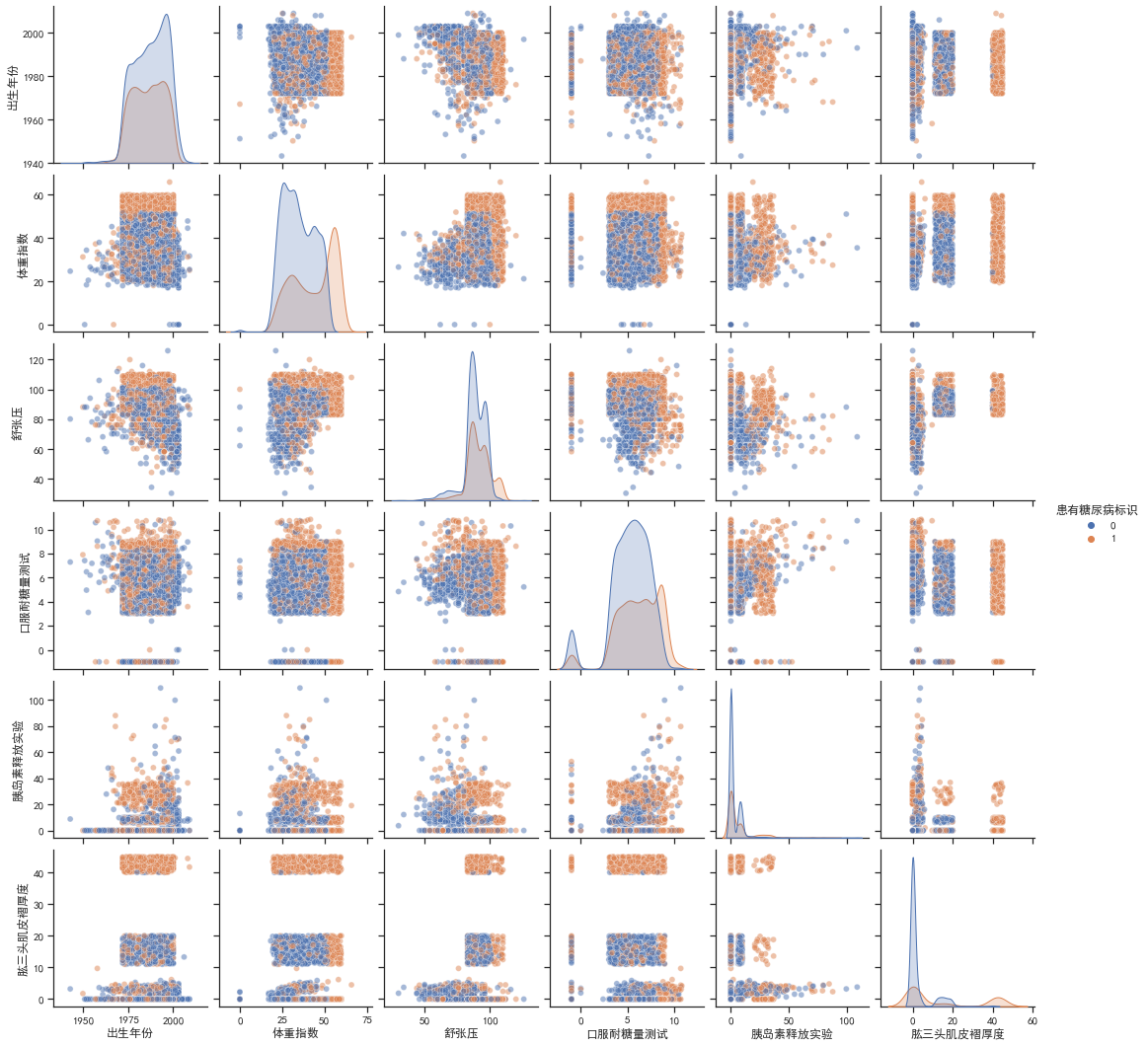
由对角线可知,患有糖尿病和非患者的体重指数和肱三头肌褶厚度的分布有较大差别,患有糖尿病的分布都略向右偏移。由其他子图可知,患者的一些联合分布特征聚集比较紧密,有一些数据还有明显的中断、不连续,可以将其作为类别特征处理,如肱三头肌褶厚度,如果在40以上就极有可能是患者,这样处理可以提高准确率。
任务3:逻辑回归尝试
- 步骤1:导入sklearn中的逻辑回归;
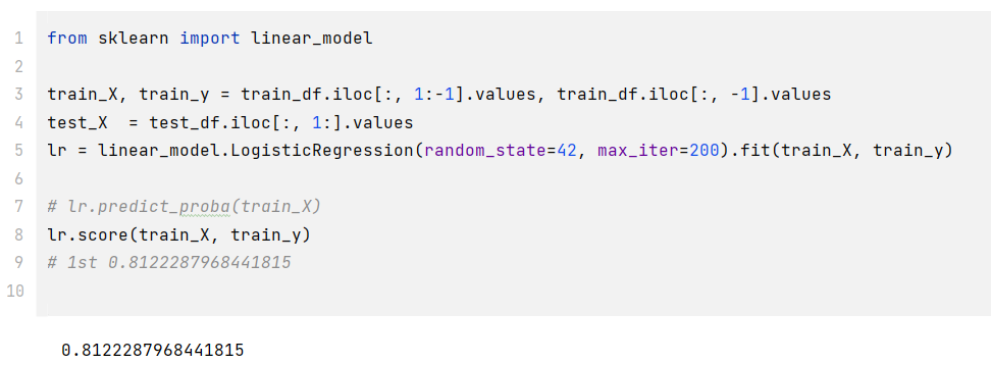
- 步骤2:使用训练集和逻辑回归进行训练,并在测试集上进行预测;
1 | |
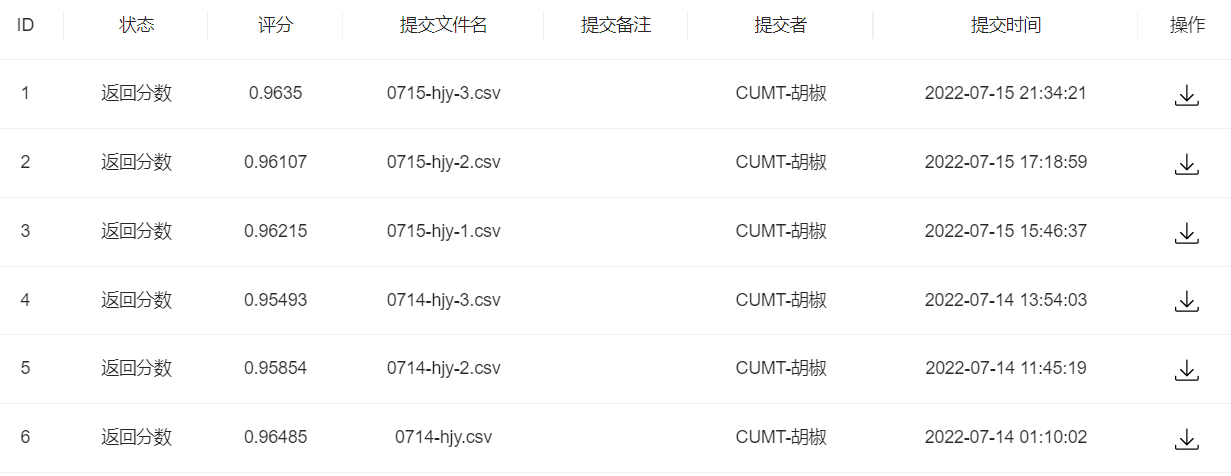
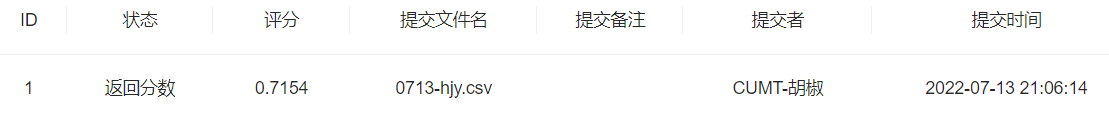
- 步骤3:将步骤2预测的结果文件提交到比赛,截图分数
1 | |
- 步骤4:将训练集20%划分为验证集,在训练部分进行训练,在测试部分进行预测,调节逻辑回归的超参数
1 | |
- 步骤5:如果精度有提高,则重复步骤2和步骤3;如果没有提高,可以尝试树模型,重复步骤2、3
1 | |
提交一波
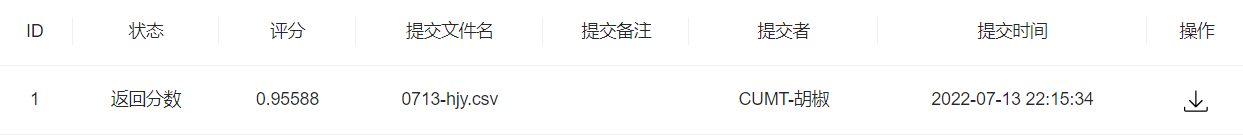
任务4:特征工程
- 步骤1:统计每个性别对应的【体重指数】、【舒张压】平均值(后面重做的时候最后发现体重指数有等于0的情况,实际上没有意义,应该设置为中位数或者平均数比较合适)
1 | |

- 步骤2:计算每个患者与每个性别平均值的差异
1 | |
- 步骤3:在上述基础上将训练集20%划分为验证集,使用逻辑回归完成训练,精度是否有提高?
1 | |
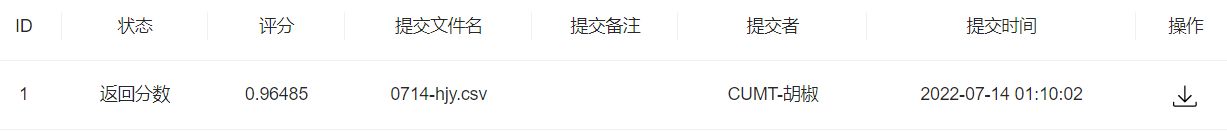
- 步骤4:思考字段含义,尝试新的特征
1 | |
任务5:特征筛选
- 步骤1:使用树模型完成模型的训练,通过特征重要性筛选出Top5的特征;
1 | |
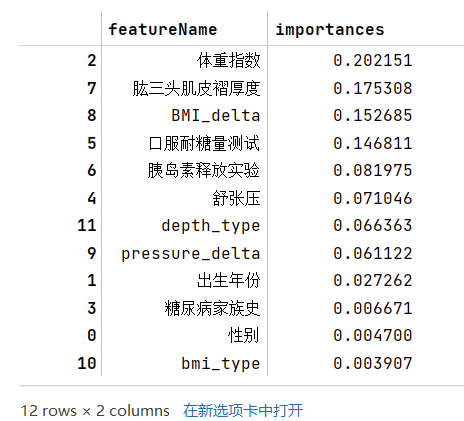
- 步骤2:使用筛选出的特征和逻辑回归进行训练,在验证集精度是否有提高?
1 | |
步骤3:如果有提高,为什么?如果没有提高,为什么?
没有提高,其他特征也比较重要,筛选的特征太少了导致细节丢失,换几个主要特征后,基本回升到原来的状态,应该是到了随机森林模型极限了,修改随机森林的参数也没有太大提升
1 | |
任务6:高阶树模型
- 步骤1:安装LightGBM,并学习基础的使用方法
- 步骤2:将训练集20%划分为验证集,使用LightGBM完成训练,精度是否有提高?
1 | |
- 步骤3:将步骤2预测的结果文件提交到比赛,截图分数,没有前面的好

- 步骤4:尝试调节搜索LightGBM的参数
1 | |
- 步骤5:将步骤4调参之后的模型从新训练,将最新预测的结果文件提交到比赛,寄

任务7:多折训练与集成
- 步骤1:使用KFold完成数据划分
1 | |
- 步骤2:使用StratifiedKFold完成数据划分
1 | |
- 步骤3:使用StratifiedKFold配合LightGBM完成模型的训练和预测(如上)
- 步骤4:在步骤3训练得到了多少个模型,对测试集多次预测,将最新预测的结果文件提交到比赛,截图分数,分数没有增长就不截图了
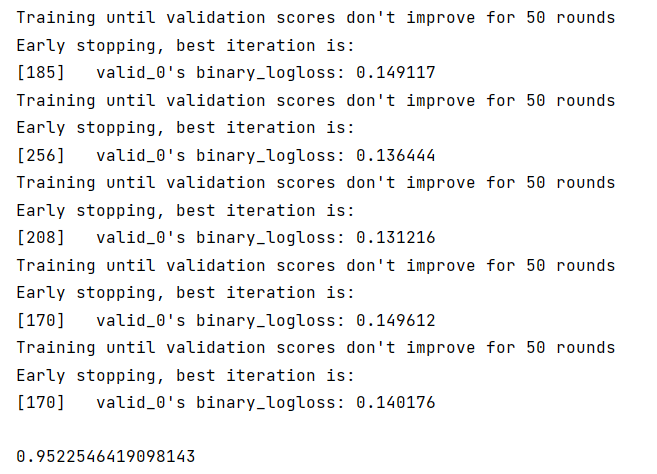
- 步骤5:使用交叉验证训练5个机器学习模型(svm、lr等),使用stacking完成集成,将最新预测的结果文件提交到比赛,截图分数
1 | |
改
1 | |
蚌埠住了,集成学习了之后还没不集成和不特征工程的分数高,傻了。